Research Summary

We use a "learn-from-nature" evolutionarily integrated strategy, Mollusks to Medicine, to discover novel peptides from venomous marine snails that could be used to manipulate cellular physiology pertaining to pain and cancer. Research projects in our lab applies inventive tools from chemistry and biology to: (1) investigate the evolution of venom in predatory marine snails, (2) discover disulfide-rich peptides from a venom source, (3) develop high-throughput methods for characterizing structure-function peptide interactions, and (4) deliver novel peptides to their site of action for therapeutic application.
Our publications highlight our peptide drug discovery strategy, demonstrate a novel computational method for optimizing the activity of venom peptide/channel complexes, provide proof in principle for the application of nanocontainers for peptide drug delivery, and our exploration of chemical pigmentation in nature. The Holford Lab’s research program is global, interdisciplinary and collaborative with impacts ranging from evolution and molecular systematics to nanotechnology, biomedicine and drug discovery.
Research Topics

Taxonomy, Phylogeny, and Systematics of Venomous Marine Snails
The Tererbridae (auger snails) are a family of globally distributed predatory marine snails that use venom to subdue their prey. The >400 described terebrid species are highly representative of a broad range of feeding strategies, more so than any other venomous marine taxa. We describe the taxonomy, phylogeny, and venom diversity of terebrid snails. Our results produced the first molecular phylogeny of the Terebridae and used it to identify terebrid lineages that used a venom apparatus similar to cone snails to produce bioactive venom peptides (teretoxins). The correlation between phylogeny and anatomical analyses is a road map for studying terebrid snails and their venom peptides. We are using terebrid taxonomy and teretoxins to clarify conoidean venom evolution.
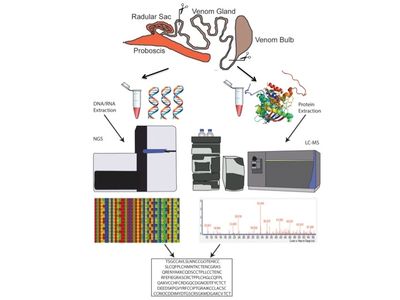
Venomics - Venom Peptide Discovery, Diversification and Evolution
Animal venoms are complex natural secretions made up of a concoction of bioactive compounds. Despite their complexity, there are generally high levels of venom convergence throughout the animal kingdom that includes similarities in gene structure and targets. We investigate questions pertaining to the evolution, production and function of venom peptides in predatory marine snails of the Terebridae. Venom peptides from predatory organisms are a resource for investigating evolutionary processes such as adaptive radiation or diversification, and exemplify promising targets for biomedical drug development. Terebridae are an understudied lineage of conoidean snails, which also includes cone snails and turrids. Characterization of cone snail venom peptides, conotoxins, has revealed a venom arsenal of bioactive peptides used to investigate physiological cellular function, predator-prey interactions, and to develop novel therapeutics. However, venom diversity of other conoidean snails remains poorly understood. My group applies a systems biology approach, venomics, which combines genomics, transcriptomics, and proteomics to discover new bioactive venom peptides and to study venom diversification in terebrids within an evolutionary phylogenetic context. Using venomics we produced the first structural characterization of a terebrid peptide, Tv1, and was the first to identify novel teretoxin venom gene superfamilies.

High Throughput Venom Peptide Characterization for Developing Novel Therapeutics
Venom peptides are levers that manipulate cell signaling. We are interested in finding new therapies for pain and cancer by creating high throughput methods for screening novel venom peptides. Each venomous marine snail of the family Conoidea can produce hundreds of novel peptides in their venom arsenal. From the breakthrough success of the first commercial venom snail drug Ziconotide (Prialt ®) used to treat chronic pain in HIV and cancer patients, we know that these peptides can potentially be used to develop novel therapies for treating human disorders. It is estimated that 15-30% of all animals on earth are venomous. This leaves a large percentage of venom arsenals understudied and uncharacterized. Collaboratively we are employing structure prediction tools (AlphaFold and trRosettaX) to develop structure prediction pipelines, peptide optimization with homology modelling and docking, peptide microarrays, microfluidic techniques, and in vitro and in vivo cell culture and whole animal assays to rapidly characterize and screen new venom peptides.

Venom Peptide Optimization and Drug Delivery
Peptides are promising therapeutic agents, however these natural compounds often need to be optimized to be used successfully as drug compounds. For example, the first snail drug approved for biomedical use, ziconotide (Prialt ®), is used to treat chronic pain in HIV and cancer patients. Despite being a breakthrough drug, ziconotide has a major drawback in that as a peptide drug, its size and complexity prevents its widespread application and a spinal tap is required to deliver it to patients. To address this issue, we are combining chemical and recombinant biology techniques to devise a method for encapsulating venom peptides from marine snails in viral capsids for delivery to their site of action. In collaborative projects we’re also developing new computational methods for optimizing the function of venom peptides to make them more selective for their molecular targets. The large size of venom peptides and the lack of solved structures of venom peptides bound to their peptide channel targets are barriers to computational docking and design of venom peptides to screen their functional activity. To address these issues, the first tool we’ve developed is ToxDock and it is freely available at http://rosie.rosettacommons.org/tox_dock.

Immune-related Micropeptide Discovery and Characterization
Similar to venom evolution, the dynamic arms-race interactions between host and pathogen contribute over time to the evolutionary selection of new immune-related genes. We are interested in exploring this evolution of new micropeptides as a response to these selective pressures. How has the evolution of these micropeptides differed across different species? Are there shared biochemical or functional characteristics of these newly evolved micropeptides? Using a combination of evolutionary, biochemical and molecular techniques, we aim to deepen our understanding of the origin and function of existing and novel immune-related micropeptides across a diversity of taxa.

Building a Better STEM World
I strongly believe that representation matters. “See it be it” is a short-term catch phrase for STEM inclusion and advancing leadership opportunities for underrepresented groups. In our initial foray in this area we wanted to generate a landscape view of the barriers and opportunities. Our first project is a systematic review of the science education literature with the aim of evaluating how and to what extent informal learning experiences impact participants awareness, interest, and engagement in STEM careers across demographic groups.
Check Out Fish Hunting Killer Snails in This Video: Striatus Bullatus